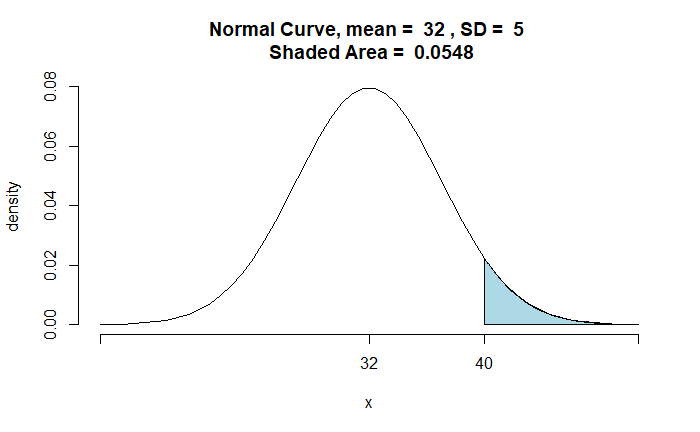
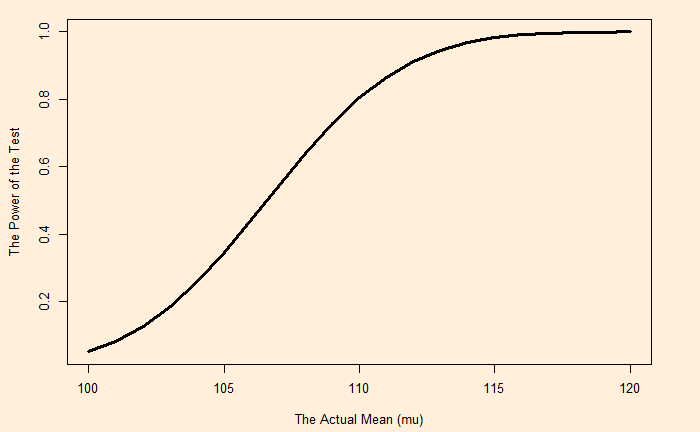
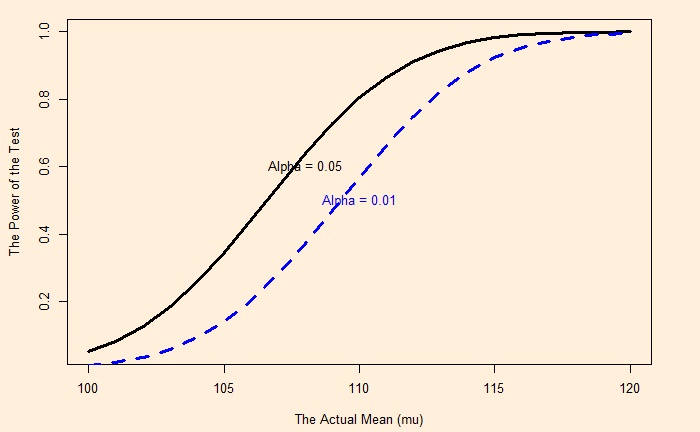
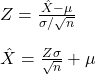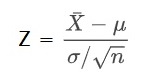Genetic Drift
We have seen natural selection as one mechanism of evolution. To clarify the whole process, this way of evolution involves two steps: 1) a random (accidental) modification of genes (mutation), followed by 2) a proliferation of a certain kind because it somehow fits well with its environment (natural selection).
But natural selection is not the only mechanism for evolution. Another means of evolution is genetic drift. In such cases, there could be no features of the genetically different versions of the species that have advantages or disadvantages from the environment. Still, random fluctuation causes one of the gene types (allele) to reduce its frequency. This also suggests that this feature is especially significant in small, isolated populations.
To give a (silly) example to distinguish the two types of evolution, we have seen the story of A moth named Biston betularia earlier. The survival probability of the black-coloured moths was higher in the industrialised (coal-polluted!) England. This is an example of natural selection. Now, think about a small population of 20 moths – 15 white and five yellows in a bright, normal neighbourhood. Someone accidentally stepped over the group, perishing seven whites and all (5) yellows. We know neither species had a special trait to withstand the boots. Yet, in the end, only eight whites were the only survivors. They then increased to a large group of white moths.