Employee Attrition Dataset
We continue using the ‘tigerstats’ package to analyse the ‘IBM HR Analytics Employee Attrition & Performance’ dataset, a fictional data set created by IBM data scientists and taken from Kaggle. The dataset contains various parameters related to attribution,
Em_data <- read.csv("D:/04Compute/Web/Employee-Attrition.csv")
str(Em_data)'data.frame': 1470 obs. of 35 variables:
$ Age : int 41 49 37 33 27 32 59 30 38 36 ...
$ Attrition : chr "Yes" "No" "Yes" "No" ...
$ BusinessTravel : chr "Travel_Rarely" "Travel_Frequently" "Travel_Rarely" "Travel_Frequently" ...
$ DailyRate : int 1102 279 1373 1392 591 1005 1324 1358 216 1299 ...
$ Department : chr "Sales" "Research & Development" "Research & Development" "Research & Development" ...
$ DistanceFromHome : int 1 8 2 3 2 2 3 24 23 27 ...
$ Education : int 2 1 2 4 1 2 3 1 3 3 ...
$ EducationField : chr "Life Sciences" "Life Sciences" "Other" "Life Sciences" ...
$ EmployeeCount : int 1 1 1 1 1 1 1 1 1 1 ...
$ EmployeeNumber : int 1 2 4 5 7 8 10 11 12 13 ...
$ EnvironmentSatisfaction : int 2 3 4 4 1 4 3 4 4 3 ...
$ Gender : chr "Female" "Male" "Male" "Female" ...
$ HourlyRate : int 94 61 92 56 40 79 81 67 44 94 ...
$ JobInvolvement : int 3 2 2 3 3 3 4 3 2 3 ...
$ JobLevel : int 2 2 1 1 1 1 1 1 3 2 ...
$ JobRole : chr "Sales Executive" "Research Scientist" "Laboratory Technician" "Research Scientist" ...
$ JobSatisfaction : int 4 2 3 3 2 4 1 3 3 3 ...
$ MaritalStatus : chr "Single" "Married" "Single" "Married" ...
$ MonthlyIncome : int 5993 5130 2090 2909 3468 3068 2670 2693 9526 5237 ...
$ MonthlyRate : int 19479 24907 2396 23159 16632 11864 9964 13335 8787 16577 ...
$ NumCompaniesWorked : int 8 1 6 1 9 0 4 1 0 6 ...
$ Over18 : chr "Y" "Y" "Y" "Y" ...
$ OverTime : chr "Yes" "No" "Yes" "Yes" ...
$ PercentSalaryHike : int 11 23 15 11 12 13 20 22 21 13 ...
$ PerformanceRating : int 3 4 3 3 3 3 4 4 4 3 ...
$ RelationshipSatisfaction: int 1 4 2 3 4 3 1 2 2 2 ...
$ StandardHours : int 80 80 80 80 80 80 80 80 80 80 ...
$ StockOptionLevel : int 0 1 0 0 1 0 3 1 0 2 ...
$ TotalWorkingYears : int 8 10 7 8 6 8 12 1 10 17 ...
$ TrainingTimesLastYear : int 0 3 3 3 3 2 3 2 2 3 ...
$ WorkLifeBalance : int 1 3 3 3 3 2 2 3 3 2 ...
$ YearsAtCompany : int 6 10 0 8 2 7 1 1 9 7 ...
$ YearsInCurrentRole : int 4 7 0 7 2 7 0 0 7 7 ...
$ YearsSinceLastPromotion : int 0 1 0 3 2 3 0 0 1 7 ...
$ YearsWithCurrManager : int 5 7 0 0 2 6 0 0 8 7 ...barchartGC
barchartGC(~Gender,data=Em_data)
barchartGC(~Attrition+Gender,data=Em_data, stack = TRUE)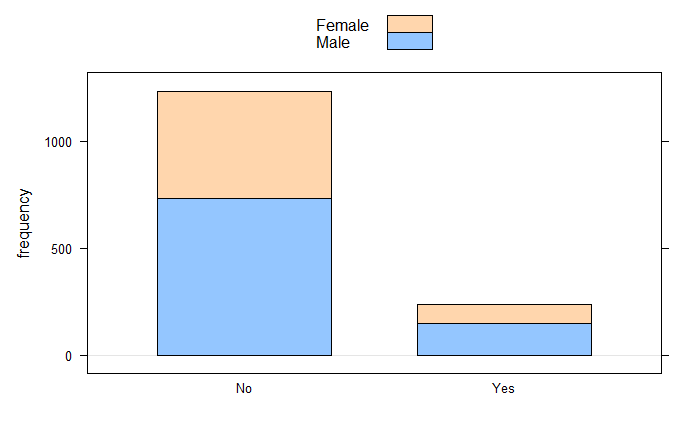
xtabs
xtabs(~Attrition+MaritalStatus,data=Em_data) MaritalStatus
Attrition Divorced Married Single
No 294 589 350
Yes 33 84 120xtabs(~Attrition+Gender,data=Em_data) Gender
Attrition Female Male
No 501 732
Yes 87 150CIMean
CIMean(~MonthlyIncome,data=Em_data)
ttestGC
ttestGC(~MonthlyIncome,data=Em_data)Inferential Procedures for One Mean mu:
Descriptive Results:
variable mean sd n
MonthlyIncome 6502.931 4707.957 1470
Inferential Results:
Estimate of mu: 6503
SE(x.bar): 122.8
95% Confidence Interval for mu:
lower.bound upper.bound
6262.062872 6743.799713 ttestGC(~Age,data=Em_data)Inferential Procedures for One Mean mu:
Descriptive Results:
variable mean sd n
Age 36.924 9.135 1470
Inferential Results:
Estimate of mu: 36.92
SE(x.bar): 0.2383
95% Confidence Interval for mu:
lower.bound upper.bound
36.456426 37.391193 Employee Attrition Dataset Read More »