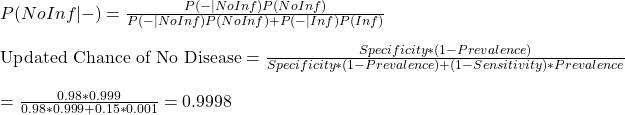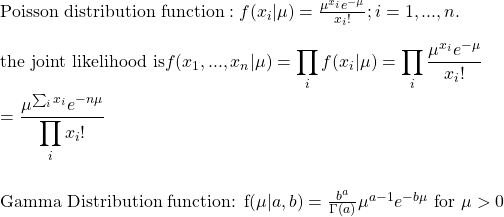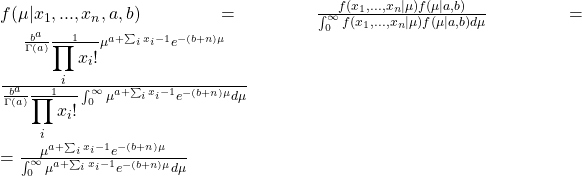Covid Stories 3 – The Gold Standard
Testing programs are not about machines but the people behind them.
We get into the calculations straight away. The equations that we made last time are:

Before we go further, let me show the output of 8 scenarios obtained by varying sensitivity and prevalence.
| Case # | Sensitivity | Specificity | Prevalence | Chance of Disease for +ve (%) | Missed in 10000 tests |
| 1 | 0.65 | 0.98 | 0.001 | 3 | 4 |
| 2 | 0.75 | 0.98 | 0.001 | 3.6 | 2.5 |
| 3 | 0.85 | 0.98 | 0.001 | 4 | 1.5 |
| 4 | 0.95 | 0.98 | 0.001 | 4.5 | 0.5 |
| 5 | 0.65 | 0.98 | 0.01 | 24 | 36 |
| 6 | 0.75 | 0.98 | 0.01 | 27 | 25 |
| 7 | 0.85 | 0.98 | 0.01 | 30 | 15 |
| 8 | 0.95 | 0.98 | 0.01 | 32 | 5 |
Note that I fixed specificity in those calculations. The leading test methods of Covid19, RT-PCR and rapid Antigen are both known to have exceptionally low false-positive rates or specificities of close to 100%.
Now the results.
Before the Spread
It is when the prevalence of the disease was at 0.001 or 0.1%. While it is pretty disheartening to know that 95% of the people who tested positive and isolated did not have the disease, you can argue that it was a small sacrifice one did for society! The scenarios of low prevalence also seem to offer a comparative advantage for carrying out random tests using more expensive higher sensitivity tests. Those are also occasions of extensive quarantine rules for the incoming crowd.
After the Spread
Once the disease has displayed its monstrous feat in the community, the focus must change from prevention to mitigation. The priority of the public health system shifts to providing quality care to the infected people, and the removal of highly infectious people comes next. Devoting more efforts to testing a large population using time-consuming and expensive methods is no more practical for medical staff, who are now required at the patient care. And by now, even the highest accurate test throws more infected people into the population than the least sensitive method when the infection rate was a tenth.
Working Smart
A community spread also rings the time to switch the mode of operation. The problem is massive, and the resources are limited. An ideal situation to intervene and innovate. But first, we need to understand the root cause of the varied sensitivity and estimate the risk of leaving out the false negative.
Reason for Low Sensitivity
The sensitivity of Covid tests is spread all over the place – from 40% to 100%. It is true for RT-PCR, even truer for rapid (antigen) tests. The reasons for an ultimate false-negative test may lie with a lower viral load of the infected person, the improper sample (swab) collection, the poor quality of the kit used, inadequate extraction of the sample at the laboratory, a substandard detector of the instrument, or all of them. You can add them up, but in the end, what matters is the concentration of viral particles in the detection chamber.
Both techniques require a minimum concentration of viral particles in the test solution. Imagine a sample that contains lower than the critical concentration. RT PCR manages this shortfall by amplifying the material in the lab, cycle by cycle, each doubling the count. That defines the cycle threshold (CT) as the number of amplification cycles required for the fluorescent signal to cross the detection threshold.
Suppose the solution requires a million particles per ml of the solution (that appears in front of the fluorescent detector), and you get there by running the cycle 21 times. You get a signal, you confirm positive and report CT = 21. If the concentration at that moment was just 100, you don’t get a response, and you continue the amplification step until you reach CT = 35 (100 x 2(35 – 21) – 2 to the power 14 – is > 1 million). The machine suddenly detects, and you report a positive at CT = 35. However, this process can’t go forever; depending on the protocols, the CT has a cut-off of 35 to 40.
On the other hand, Antigen tests detect the presence of viral protein, and it has no means to amplify the quantity. After all, it is a quick point of care test. A direct comparison with the PCR family does not make much sense, as the two techniques work on different principles. But reports suggest sensitivities of > 90% for antigen tests for CT = 28 and lower. You can spare a thought at the irony that an Antigen test is sensitive to detect the presence of the virus that the PCR machine would have taken 28 rounds of amplification. But that is not the point. If you have the facility to amplify, why not use it.
The Risk of Leaving out the Infected
It is a subject of immense debate. Some scientists argue that the objectives of the testing program should be to detect and isolate the infectious and not every infected. While this makes sense in principle, there is a vital flaw in the argument. There is an underlying assumption that the person with too few counts to detect is always on the right side of the infection timeline – in the post-infectious phase. In reality, the person who got the negative test in a rapid screening can also be in the incubation period and becomes infectious in a few days. They point to the shape of the infection curve, which is skewed to the right, or fewer days to incubate to sizeable viral quantity and more time on the right. Another suggestion is to test more frequently so that the person who missed due to a lower count comes back for the test a day or two later and then caught.
How to Increase Sensitivity
There are a bunch of activities the system can do. The first in the list is to tighten the quality control or prevent all the loss mechanisms from the time of sampling till detection. That is training and procedures. The second is to change the strategy from analytical regime to clinical – from random screening to targetted testing. For example, if the qualified medical professional identifies patients with flu-like symptoms, the probability of catching a high-concentrated sample increases. Once that sample goes to the testing device for the antigen, you either find the suspect (covid) or not (flu), but it was not due to any lack of virus from the swab. If the health practitioner still suspects, she may recommend an RT PCR, but no more a random decision.
In Summary
We are in the middle of a pandemic. The old ways of prevention are no more practical. Covid diagnostics started as a clinical challenge, but somewhere along the journey, that shifted more to analytics. While test-kit manufacturers, laboratories, data scientists and the public are all valuable players to maximise the output, the lead must go back to trained medical professionals. A triage system, based on experiences to identify symptoms and suggested follow up actions, is a strategy worth the effort to stop this deluge of cases.
Further Reading
Interpreting a Covid19 test result: BMJ
Issues affecting results: Exp Rev Mol Dia
False Negative: NEJM
Rapid Tests – Guide for the perplexed: Nature
Real-life clinical sensitivity of SARS-CoV-2 RT-PCR: PLoS One
Diagnostic accuracy of rapid antigen tests: Int J Infect Dis
Rapid tests: Nature
Rethinking Covid-19 Test Sensitivity: NEJM
Cycle Threshold Values: Public Health Ontario
CT Values: APHL
CT Values: Public Health England
Covid Stories 3 – The Gold Standard Read More »