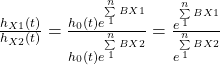The Truth that Exists in Mathematical Models
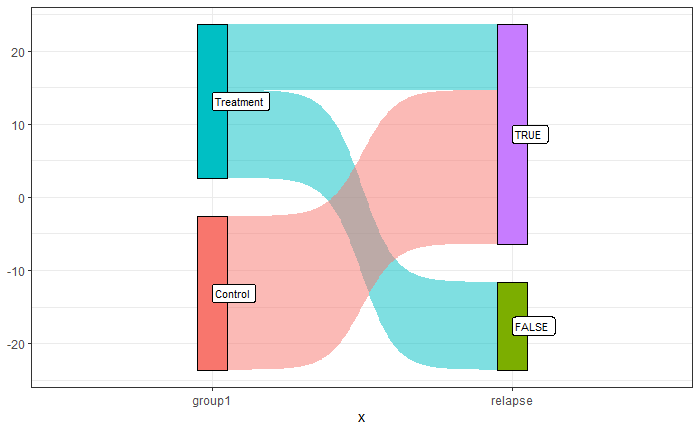
The concept of comparative advantage is something we saw when we analysed international trade as beneficial to both participating countries. Yet, the notion remains highly challenged by the common public. Trade, in their mind, remains a zero-sum game. If I go and sell goods in a foreign country, they lose, and I win.
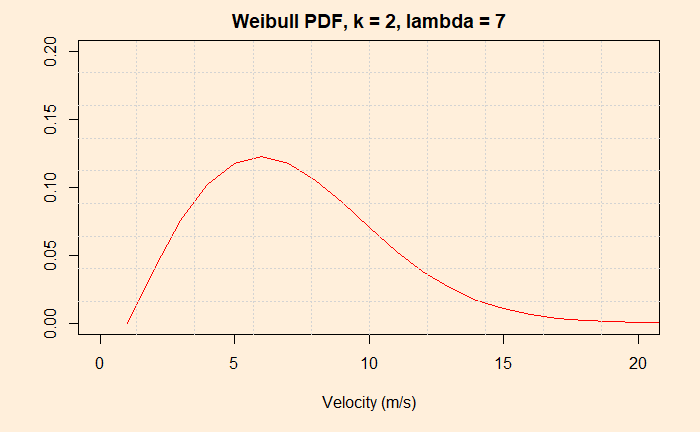
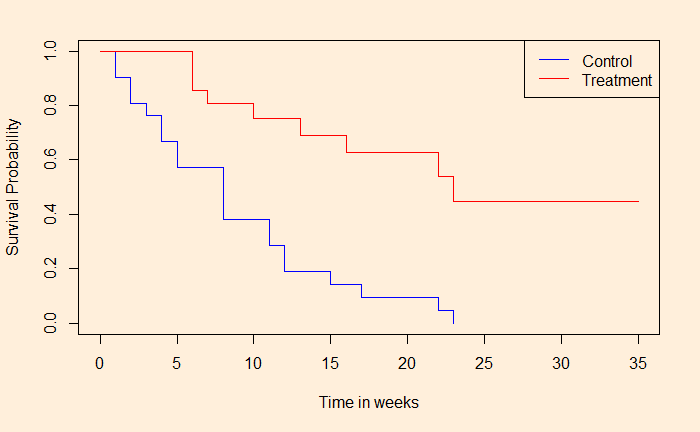
Similarly, the role that the climate models play in understanding global warming. No matter how hard one tries to prove the fact using charts and equations, the public still requires to hear stories of hardships of extreme weather events to move their views. Mathematical models are inevitable as we are dealing with a complex problem with many factors that are not related linearly to the climate.
Part of the blame for this situation falls on the economists and scientists themselves. Most often, they assume the concepts they develop are simple and intuitive, and those who don’t understand are some less intelligent type.
On the other hand, people grow up hearing exaggerated stories about common sense and simple narratives. Models and equations are too difficult to understand and, therefore, some form of a trick played by the proponent.
The Truth that Exists in Mathematical Models Read More »