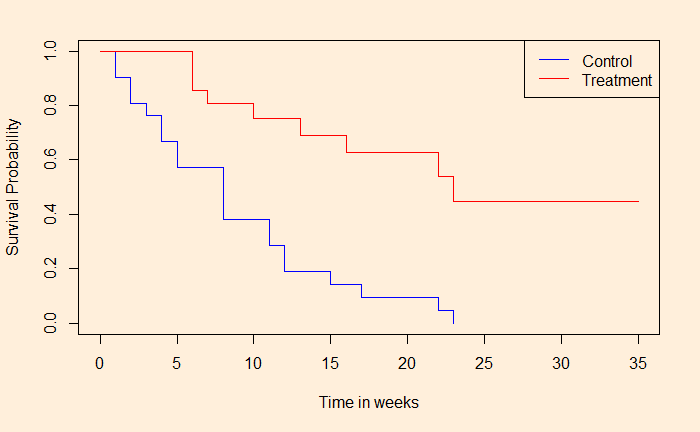
Here is where we stopped last time. The next step is quantifying the difference between the treatment and the control groups. Now refresh your memory or hazard ratio, efficacy – all those stuff.
Cox proportional hazards model
The main idea is to find out if the survival time depends on one or more variables or predictors. In our case, there is only one variable with two values – treatment or placebo. Cox model does regression (curve fitting or history matching) of the survival curve using the predictor. The model has an exponential relationship between the observed hazard to the effect of the predictor.
![]()
h(t) is the observed hazard (a function of time), and h0(t) is the baseline hazard. The exponential term is the effect of the condition (treatment or not). Note that the exponential term is not a function of time, and eB1 is the hazard ratio. We know we have two conditions for X1, i.e. X1 = 1 (treatment) and X1 = 0 (control).
![]()
The above is the ratio between the hazard when the treatment is present and the hazard when the treatment is absent.
Note the regression can be performed using a combination of variables, e.g. age, sex etc.
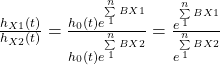
Significance
The following R commands do all the job and spit out the hazard ratio and the significance or p-value.
ill_cox_fit <- coxph(Surv(weeks, illness) ~ group, data = ill_data1)The output is:
Call:
coxph(formula = Surv(weeks, illness) ~ group, data = ill_data1)
n= 42, number of events= 30
coef exp(coef) se(coef) z Pr(>|z|)
groupTreatment -1.5721 0.2076 0.4124 -3.812 0.000138 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
exp(coef) exp(-coef) lower .95 upper .95
groupTreatment 0.2076 4.817 0.09251 0.4659
Concordance= 0.69 (se = 0.041 )
Likelihood ratio test= 16.35 on 1 df, p=5e-05
Wald test = 14.53 on 1 df, p=1e-04
Score (logrank) test = 17.25 on 1 df, p=3e-05The ‘exp(coef)’ value is nothing but the hazard ratio. And we know that the efficacy is (1 – hazard ratio), and in our case, it is about 80%. The p-value is low, and therefore the difference in survival time between the treatment and control is statistically significant.