Let’s do a logistic regression of health data. Experiments with the Cleveland database focused on distinguishing the presence (value: 1,2,3,4) from the absence (value 0). The featured health parameters are
Age
Sex
CP: chest pain
Trestbps: resting blood pressure (mm Hg)
Chol: serum cholesterol (mg/dl)
Fbs: fasting blood sugar > 120 mg/dl
Restecg: Rest ECG
Thalach: maximum heart rate achieved during the thallium stress test
Exang: exercise-induced angina
Oldpeak: ST depression induced by exercise relative to rest
Slope: the slope of the peak exercise ST segment
Ca: number of major vessels (0-3) coloured by fluoroscopy
Thal:
Hd: diagnosis of heart disease
After cleaning up and conditioning, the data looks like this:
297 obs. of 14 variables:
$ Age : num 63 67 67 37 41 56 62 57 63 53 ...
$ Sex : Factor w/ 2 levels "F","M": 2 2 2 2 1 2 1 1 2 2 ...
$ CP : Factor w/ 4 levels "1","2","3","4": 1 4 4 3 2 2 4 4 4 4 ...
$ Trestbps: num 145 160 120 130 130 120 140 120 130 140 ...
$ Chol : num 233 286 229 250 204 236 268 354 254 203 ...
$ Fbs : Factor w/ 2 levels "0","1": 2 1 1 1 1 1 1 1 1 2 ...
$ Restecg : Factor w/ 3 levels "0","1","2": 3 3 3 1 3 1 3 1 3 3 ...
$ Thalach : num 150 108 129 187 172 178 160 163 147 155 ...
$ Exang : Factor w/ 2 levels "0","1": 1 2 2 1 1 1 1 2 1 2 ...
$ Oldpeak : num 2.3 1.5 2.6 3.5 1.4 0.8 3.6 0.6 1.4 3.1 ...
$ Slope : Factor w/ 3 levels "1","2","3": 3 2 2 3 1 1 3 1 2 3 ...
$ Ca : Factor w/ 4 levels "0","1","2","3": 1 4 3 1 1 1 3 1 2 1 ...
$ Thal : Factor w/ 3 levels "3","6","7": 2 1 3 1 1 1 1 1 3 3 ...
$ Hd : Factor w/ 5 levels "0","1","2","3",..: 1 3 2 1 1 1 4 1 3 2 ...logic <- glm(Hd ~ ., data = heart_data, family = "binomial")
predicted.data <- data.frame(Prob.HD = logic$fitted.values, HD = heart_data$Hd)
par(cex=0.8, mai=c(0.7,0.7,0.2,0.5), bg = "antiquewhite1")
plot(x = predicted.data$HD, y = predicted.data$Prob.HD)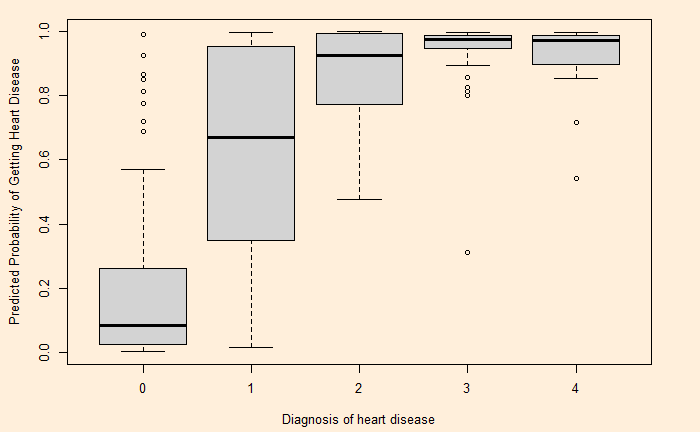
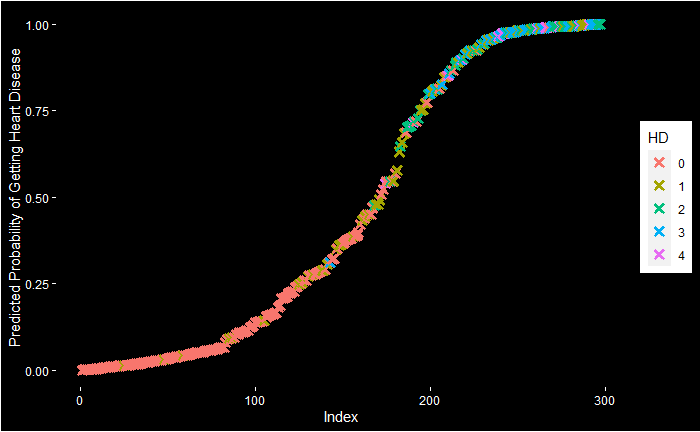
An even fancier plot can be made using the following code:
logic <- glm(Hd ~ ., data = heart_data, family = "binomial")
predicted.data <- data.frame(Prob.HD = logic$fitted.values, HD = heart_data$Hd)
predicted.data <- predicted.data[order(predicted.data$Prob.HD, decreasing = FALSE),]
predicted.data$Rank <- 1:nrow(predicted.data)
ggplot(data = predicted.data, aes(x = Rank, y = Prob.HD)) +
geom_point(aes(color = HD), alpha = 1, shape = 4, stroke = 2) +
xlab("Index") +
ylab("Predicted Probability of Getting Heart Disease") +
theme(text = element_text(color = "white"),
panel.background = element_rect(fill = "black"),
plot.background = element_rect(fill = "black"),
panel.grid = element_blank(),
legend.text = element_text(color = "black"),
legend.title = element_text(color = "black"),
axis.text = element_text(color = "white"),
axis.ticks = element_line(color = "white"))